Data Dictionary¶
This document includes the details of every table and field in the
database. The details of possible data types are described in
Data Types.
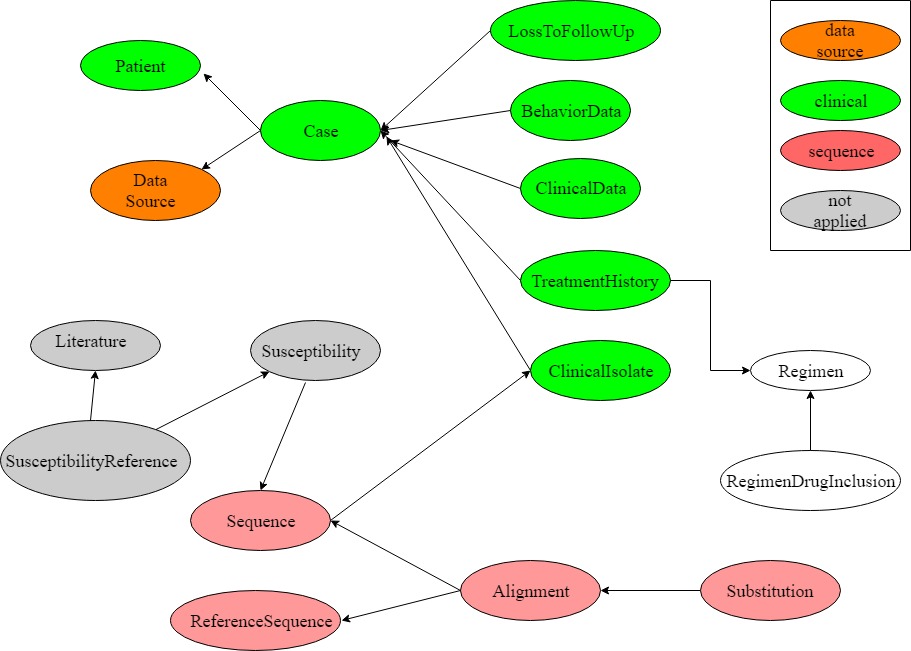
Relationships between tables in the SHARED schema.
Patient¶
A study participant
| Req’d | Name | Data Type | Description |
|---|---|---|---|
|
|
patient_id |
uuid |
Unique identifier |
sex |
enum(male, female, other) |
The participant’s sex at birth | |
ethnicity |
enum(am-nat, asian, black, hisp, pa-isl, white) |
The participant’s ethnicity: American Native, Asian, Black/African American, Hispanic/Latino, Pacific Islander, or White | |
year_of_birth |
integer |
Participant’s year of birth (e.g: 1985) |
Data Source¶
A collaborating site (provides participant data)
| Req’d | Name | Data Type | Description |
|---|---|---|---|
|
|
source_id |
uuid |
Unique identifier |
|
|
collaborator_name |
string |
Name of the collaborator |
country |
VARCHAR(2) |
ISO 3166-1 alpha-2 code | |
submission year |
Integer |
submission year | |
data merger |
Integer |
| Req’d | Name | Data Type | Description |
|---|---|---|---|
|
|
case_id |
uuid |
Unique identifier |
|
|
patient_id |
foreign key (Patient) |
The person who contributed the data in this case. |
source_id |
foreign key (DataSource) |
The study the participant entered the database with | |
country |
string |
The country where the person participated in the study (as an ISO 3166-1 alpha-2 code) | |
study_participant_id |
string |
The unique identifier that the source study gave to the person contributing this data | |
data_modified |
bool |
Has the data been modified? | |
upload_date |
date |
date when the data is uploaded |
LossToFollowUp¶
Records data about participants leaving the study
| Req’d | Name | Data Type | Description |
|---|---|---|---|
|
|
ltfu_id |
uuid |
unique identifier |
|
|
case_id |
foreign key (Case) |
A collection of related data from a study participant |
ltfu_year |
integer |
The year the participant was lost to follow-up (e.g: 2012) | |
died |
bool |
Is the participant deceased? | |
cod |
enum(liv, aid, odo, can, cir, res, dia, gen, tra, cer, dig, oth) |
Cause of death (if applicable; blank otherwise) |
BehaviorData¶
Behavioral information about a participant
| Req’d | Name | Data Type | Description |
|---|---|---|---|
|
|
behaviordata_id |
uuid |
Unique identifier |
|
|
case_id |
foreign key (Case) |
A collection of related data from a study participant |
sex_ori |
enum (heterosexual, homosexual, bisexual, other) |
Participant’s sexual orientation | |
idu |
bool |
Injection drug use? (ever) | |
ndu |
bool |
Non-injection drug use? (ever) | |
idu_recent |
bool |
Injection drug use in the past 6 months? | |
ndu_recent |
bool |
Non-injection drug use in the past 6 months? | |
prison |
bool |
Has the participant been in prison (ever)? |
ClinicalData¶
Participants’ test results and relevant medical history
| Req’d | Name | Data Type | Description |
|---|---|---|---|
|
|
clinicaldata_id |
uuid |
Unique identifier |
case_id |
foreign key(Case) |
A collection of related data from a study participant | |
clinical_kind |
string (bl, eot,fw4 .etc) |
Is this clinical data from baseline or follow-up? | |
hiv |
bool |
Is the participant co-infected with HIV? | |
hbv |
bool |
Is the participant co-infected with HBV? | |
vl |
float |
Viral Load (in IU/mL) | |
ost |
bool |
Has the participant undergone opioid substitution therapy in the last six months? | |
cirr |
bool |
Does the participant have cirrhosis? | |
fibrosis |
enum(F0, F0-F1, F1, F2, F3, F4) |
METAVIR fibrosis score | |
inflammation |
enum(A0, A1, A2, A3) |
METAVIR inflammation score | |
metavir_by |
enum(fibroscan, biopsy, clinical, other) |
Method used to determine metavir score: | |
stiff |
float |
Liver stiffness (in kPa). | |
alt |
float |
Alanine aminotransferase level (in U/L). | |
ast |
float |
Aspartate aminotransferase level (in U/L). | |
crt |
float |
Creatinine level (in mg/dL). | |
egfr |
float |
Estimated glomerular filtration rate (in mL/min). | |
ctp |
float |
Child-Turcotte-Pugh score. | |
meld |
float |
MELD score. | |
ishak |
float |
Ishak score. | |
bil |
float |
Bilirubin test result, in mg/dL. | |
hemo |
float |
Hemoglobin test result, in g/dL. | |
alb |
float |
Albumin test result, in g/dL. | |
inr |
float |
International Normalized Ratio test result. | |
phos |
float |
Phosphate test result, in mg/dL. | |
urea |
float |
Urea test result, in ng/dL. | |
plate |
float |
Platelet count, in cells/mm3. | |
CD4 |
float |
CD4 count, in cells/mm3. | |
crp |
float |
C-Reactive Protein test result, in mg/L. | |
il28b |
enum(TT,CT,CC) |
The participant’s IL28B-rs12979860 genotype: TT, CT, CC | |
asc |
bool |
Did the participant have ascites before or during treatment? | |
var_bleed |
bool |
Did the participant have variceal bleeding before or during treatment? | |
hep_car |
bool |
Did the participant have hepatocellular carcinoma before or during treatment? | |
transpl |
bool |
Has the participant had a liver transplant? |
TreatmentHistory¶
Information about a participant’s treatment
| Req’d | Name | Data Type | Description |
|---|---|---|---|
|
|
treatmenthistory_id |
uuid |
Unique identifier |
|
|
case_id |
foreign key (Case) |
A collection of related data from a study participant |
clinical_kind |
string (bl, eot,fw4 .etc) |
Is this clinical data from baseline or follow-up? | |
treatment_status |
enum(TN, TE) |
treatment status associated with submitted sequence TN: treatment naïve(first time treatment); TE: treatment experienced |
|
duration_act |
integer |
The treatment’s actual duration (in days), if different from the scheduled duration | |
current_regimen_id |
foreign key (Regimen) |
The drug regimen taken by the participant | |
current_treatment_response |
enum(SVR, NR, EOT, BT, RL, RI, VF, DC) |
Viral response: sustained viral response, non-responsive, detectable viral load at end-of-treatment, viral-breakthrough during treatment, viral relapse, reinfection, virological failure, or discontinued | |
prev_regimen_id |
foreign key (Regimen) |
If the participant has been treated before, what is the last regimen they were on? | |
prev_regimen_duration |
integer |
Previous treatment’s actual duration (in days) | |
pprev_regimen_id |
foreign key (Regimen) |
If the participant has been treated before, what regimen were they on before-last? | |
pprev_regimen_duration |
integer |
Previous previous treatment’s actual duration (in days) |
Regimen¶
A collection of kinds and ammounts of drugs used in treatment
| Req’d | Name | Data Type | Description |
|---|---|---|---|
|
|
regimen_id |
string |
DAA components name, e.g. SOF+LDV |
name |
string |
The trade name of this treatment, if applicable, e.g. Harvoni | |
duration_planned |
integer |
planned duration of the regimen (in days) |
RegimenDrugInclusion¶
The drugs in a regimen, and their doses and durations
| Req’d | Name | Data Type | Description |
|---|---|---|---|
DAA_id |
enum(ASV, BOC, DCV, DAS, DAS_XR, EBR, GLP, GZR, LDV, NPV, OMB, PAR, PEG, PIB, RBV, RIT, SIM, SOF, TVR, VAN, VEL, VOX, other, none) |
The three letter code for a DAA component. | |
regimen_id |
foreign key(Regimen) |
DAA components name, e.g. SOF+LDV | |
dose |
float |
Dosage of the medication prescribed (in mg) | |
frequency |
enum(QD, BID, TID, QID, QWK) |
How often is this medication taken | |
duration |
integer |
planned duration of the DAA |
ClinicalIsolate¶
Isolate information
| Req’d | Name | Data Type | Description |
|---|---|---|---|
|
|
clinical_isolate_id |
uuid |
unique identifier |
|
|
case_id |
foreign key (Case) |
A collection of related data from a study participant |
seq_kind |
string (bl, eot, fw4 ,vf.etc) |
Whether this isolate is from a baseline sample, an end-of-treatment-sample, or a follow up sample 4, 12, or 24 weeks after end-of-treatment. | |
clinical_gt |
string |
genotype as determined by clinical array or by submitting collaborator | |
clinical_subtype |
string |
subtype as determined by clinical array or by submitting collaborator |
Sequence¶
Sequences and data needed for rapid alignment
| Req’d | Name | Data Type | Description |
|---|---|---|---|
|
|
seq_id |
uuid |
Unique identifier |
|
|
clinical_isolate_id |
foreign key (ClinicalIsolate) |
Clinical_isolate the sequence was obtained from |
seq_genotype |
enum(1, 2, 3, 4, 5, 6, mixed, recombinant, indeterminate) |
sequence derived genotype | |
seq_subgenotype |
string |
sequence derived sub-genotype | |
seq_method |
enum(sanger, ngs) |
The sequencing method used on this isolate | |
cutoff |
float |
The cutoff-fraction used to generate a consensus sequence; ‘5%’ is stored as ‘0.05’ | |
|
|
raw_nt_seq |
string |
The raw nucleotide in the assembled sequence |
amino_acid_seq |
string |
amino acid sequence derived from raw_nt_seq |
ReferenceSequence¶
Reference nucleotide sequences used for alignment
| Req’d | Name | Data Type | Description |
|---|---|---|---|
|
|
reference_id |
string |
The reference sequence’s genotype |
reference_strain |
string |
strain of reference sequence | |
|
|
genebank |
string |
Genbank accession number e.g. NC_004102 |
length_bp |
integer |
length | |
pos_NS3-4a_start |
integer |
start position | |
pos_NS3-4a_end |
integer |
end position | |
pos_NS5a_start |
integer |
start position | |
pos_NS5a_end |
integer |
end position | |
pos_NS5b_start |
integer |
start position | |
pos_NS5b_end |
integer |
end position | |
genome-nt-seq |
string |
Raw nucleotide sequence of whole genome | |
ns3-nt-seq |
string |
ns3 nucleotide sequence | |
ns5a-nt-seq |
string |
ns5a nucleotide sequence | |
ns5b-nt-seq |
string |
ns5b nucleotide sequence | |
ns3-aa-seq |
string |
ns3 amino acid sequence | |
ns5a-aa-seq |
string |
ns5a amino acid sequence | |
ns5baa-seq |
string |
ns5b amino acid sequence |
Alignment¶
A gene found in a sequence
| Req’d | Name | Data Type | Description |
|---|---|---|---|
|
|
alignment_id |
uuid |
Unique identifier |
|
|
seq_id |
foreign key (Sequence) |
The sequence used for alignment |
|
|
reference_id |
foreign key (ReferenceSequence) |
The reference sequence this alignment was performed with respect to |
|
|
nt_start |
integer |
The first position of the alignment in the raw nucleotide sequence |
|
|
nt_end |
integer |
The last position of the alignment in the raw nucleotide sequence |
|
|
target_gene |
string |
The name of the gene this alignment is in |
Substitution¶
A substitution, insertion, or deletion in an RNA sequence
| Req’d | Name | Data Type | Description |
|---|---|---|---|
|
|
alignment_id |
foreign key (Alignment) |
The alignment this substitution is found in |
|
|
position |
integer |
Nucleotide position (with respect to the reference sequence) |
|
|
kind |
enum (simple, insertion, deletion) |
Kind of subsitution (‘simple’ for a single nucleotide polymorphism, insertion or deletion for gaps) |
sub_aa |
string |
For a simple substitution, the new amino acid (blank for insertions and deletions) | |
insertion |
string |
For an insertion, the inserted Amino Acid sequence (blank for SNPs and deletions) | |
deletion_length |
integer |
For a deletion, the number of deleted amino acids (blank for SNPs and insertions) |
Susceptibility¶
Susceptibility test results
| Req’d | Name | Data Type | Description |
|---|---|---|---|
|
|
susceptibility_id |
uuid |
Unique id |
seq_id |
foreign key (Sequence) TODO! |
The sequence being tested | |
susc_method |
enum(luciferase, qpcr, bdna, beta-gal) |
Method used to measure susceptibility | |
medication |
enum(ASV, BOC, DCV, DAS, EBR, GLP, GZR, LDV, OMB, PAR, PEG, PIB, RBV, RIT, SIM, SOF, TVR, VAN, VEL, VOX) |
The three letter id of the medication being tested | |
result |
float |
Concentration of medication required for inhibition (in nM) | |
result_bound |
enum (<, =, >) |
||
ic |
enum (50, 90, 95) |
percent inhibition | |
fold |
float |
Fold-change compared to wild type | |
fold_bound |
enum (<, =, >) |
Represents uncertainty in the fold-change measurement |
Literature¶
A reference to a publication
| Req’d | Name | Data Type | Description |
|---|---|---|---|
ref_lit_id |
uuid |
Unique id | |
author |
string |
||
title |
string |
||
journal |
string |
||
year |
string |
Year of publication | |
url |
string |
URL to the reference online | |
publication_dt |
date |
Null for unpublished results | |
pubmed_id |
string |
SusceptibilityReference¶
Marks the reference supporting a susceptibility result
| Req’d | Name | Data Type | Description |
|---|---|---|---|
susceptibility_id |
foreign key (Susceptibility) |
The marked susceptibility | |
ref_lit_id |
foreign key (Literature) |
The associated literature reference |
